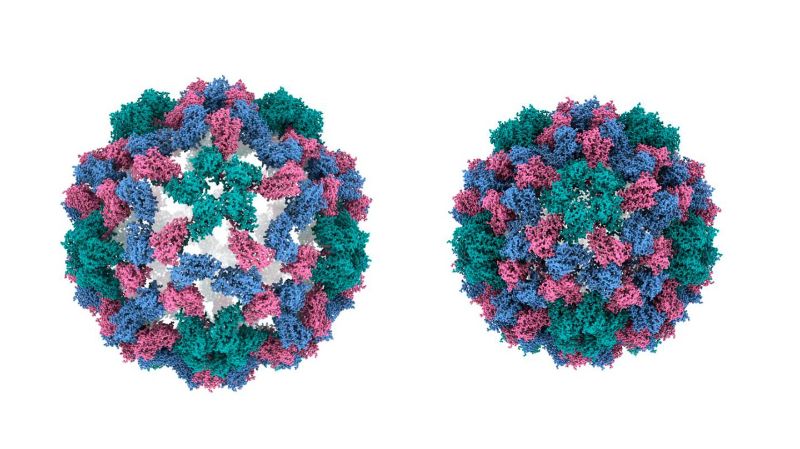New imaging technique “sees” virus move in unprecedented detail
Proteins are the workhorses of biological systems, carrying out their work with extraordinary precision and speed. For years, observing proteins in action has been a significant challenge, as imaging methods often lacked sufficient speed and resolution to capture their elegant but swift dances.
Now, a team of scientists led by Professor Ulrich Lorenz at EPFL, has used a novel imaging technique that pushes the time-resolution of cryo-electron microscopy (cryo-EM) down to microseconds, to observe the fast dynamics of a virus in real-time. The study is published in Nature Communications.
The researchers first developed the imaging technique in 2021, based on cryoEM, a technique that can capture pictures of biomolecules such as proteins with atomic precision. In cryoEM, samples are embedded in vitreous ice, a glass-like form of ice that is obtained when water is frozen so rapidly that crystallization cannot occur. With the sample vitrified, high-resolution pictures of its molecular structure can be taken with an electron microscope, an instrument that forms images using a beam of electrons instead of light.
The innovative cryoEM method won its inventors, Jacques Dubochet, Joachim Frank, and Richard Henderson the Nobel Prize in Chemistry in 2017. In 2021, Lorenz and his lab extended the capabilities of cryoEM, to capture images of protein movements at the microsecond (a millionth of a second) timescale by rapidly melting the vitrified sample with a laser pulse. As the ice melts into a liquid, it opens a “tunable” time window in which the protein can be induced to move in the way they do in their natural liquid state in the cell.
Using the same approach, the researchers have now used their technique to capture rapid viral movements with unparalleled precision. The team focused on the cowpea chlorotic mottle virus (CCMV), a plant virus known for its large-amplitude motions crucial to its infection cycle. It is known that a change in pH causes the virus's capsid (a protective shell) to expand rapidly, and using the new technique, the team was able to observe the actual mechanics of this process.
“It is an expansion of the capsid that occurs when the virus infects a cell,” says Ulrich Lorenz. “We studied this process in reverse, i.e., the contraction of the capsid, which allowed us to elucidate the mechanics of the capsid in a more straightforward manner.”
The new imaging technique worked wonders. “We got a very detailed picture of the functioning and mechanics of this nanoscale machine, which includes the surprising insight that different motions of the capsid proteins occur at different speeds,” says Lorenz. “We also learned that the contraction, even though it is a large-amplitude motion, is very rapid, with the virus in its extended state resembling a stretched spring that is suddenly released and contracts.”
Beyond the virus, the new microsecond time-resolved cryo-EM technique addresses the broader challenge of observing proteins as they function. “We show, for the first time, that our method can be used to observe a process that actually occurs in nature,” says Lorenz. “No other method exists that would be able to make this type of observation. If it becomes possible to extend our experiments to a broad range of systems, which we firmly believe is the case, our method has the potential to revolutionize our understanding of how proteins function.”